|
|
Banner Art: DNA strand bridged by abacus, symbolizing the extensive collaboration between biologists and computer scientists in shaping the genomics revolution. Credit: Prof. Ergun Akleman, Texas A&M Visualization Department (crafted specifically for the paper Petersen et al., Sci. Adv. 2018;4: eaat4211) |
|
|
Cross-Disciplinary Work Key Factor in Genomics Revolution Research Finds
How and Why Gene Hunting Changed the Culture of Science
HOUSTON, August 2018 – Years after the end of the Human Genome Project (HGP), which mapped the human genetic blueprint, the ramifications to science and its culture are still unfolding.
Ioannis Pavlidis, Eckhard Pfeiffer Professor of Computational Physiology at the University of Houston, Alexander Petersen, Professor of Management at UC, Merced, and their colleagues are reporting
in Science Advances that HGP scientists not only laid the groundwork for scientific breakthroughs for decades to come, but also brought to the mainstream a collaboration model that
changed science’s cultural norms.
“One of the key factors of the success was the way it incorporated cross collaboration between biologists, computer scientists and other disciplines,” said Pavlidis. “Research was organized around a new model, the consortium model, where scientists from different fields and in different localities worked for years towards a common goal.”
Since Galileo first peered into a telescope and DaVinci sketched out human anatomy, the business of science has been populated by independent endeavors. Systematic team efforts were transient exceptions. For centuries the image of the loner scientist was not updated, nor did it need to be – until recently. Pavlidis and Petersen report that after the sequencing of the human genome was completed in 2003, the consortium model did not go away, and scientists never returned to their silos. In a clear departure from norms, new and old researchers from biology, computing, and other disciplines kept building consortia, unlocking the genetic secrets of fauna and flora, and in the process transforming our ability to understand, predict, and edit life.
“In an era where science has been professionalized, one cannot fully appreciate the reasons for genomics’ success unless s/he looks at the effect it had on the researchers’ careers,” said Petersen.
To investigate this, the authors assembled a dataset that included biographic and funding information for over 4,000 faculty in 155 biology and computing departments in the United States, including information for their 400,000+ publications co-authored with a network of nearly 500,000 collaborators.
Using big data and counterfactual modeling, the team demonstrated that biology and computing researchers who crossed disciplinary boundaries collaborating with each other on genomics grounds had more successful careers than their mono-disciplinary colleagues.
“Their cross-disciplinary publications had superior impact and the same patterns were found to be true at the international literature level,” said Petersen. Specifically, genomic articles with mixed authorship from biology and computing, or genomic articles with mixed biology and computing content, had significantly stronger impact compared to alternatives.
Petersen and Pavlidis believe that as cross-disciplinary work continues, scientific advancements will follow suit. “If we continue to do this and have well-timed and well-designed funding efforts, as the Human Genome Project had, then I think we can put scientific progress on steroids,” said Pavlidis.

|
|
|
|
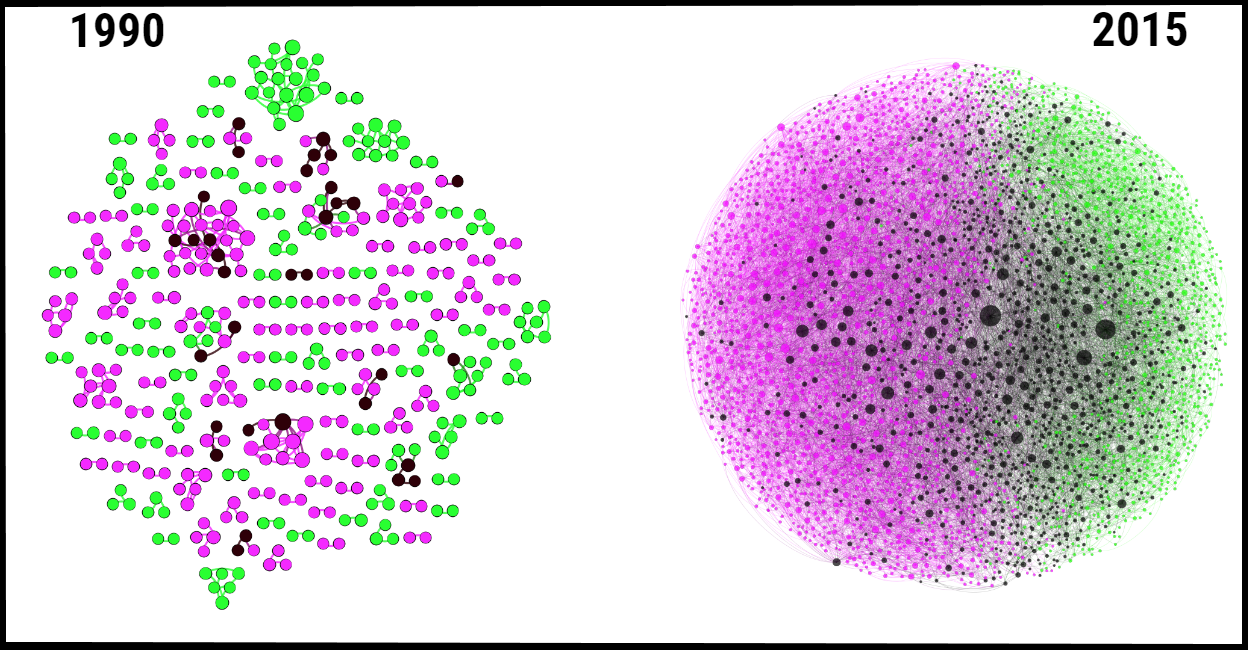
FIGURE 1: Cross-disciplinarity key factor in genomics revolution Evolution of the collaboration network in the biology-computing college in the United States from 1990 (pre Human Genome Project era) to 2015. Links represent co-authorship relationships. Green nodes denote biology faculty, magenta nodes denote computing faculty, and black nodes denote faculty from either discipline involved in cross-disciplinary genomics work. The size of the nodes suggest the centrality of the faculty in the network. 1990: the collaboration among faculty in the biology-computing college in the United States is characterized by few, small-size groups, working in isolation. 2015: the biology-computing college in the United States has grown into a massive interconnected network, where genomics faculty constitute a prominent part. Credit: The figure was generated by the network model reported by Petersen et al., Sci. Adv. 2018;4: eaat4211 |
|
|
|
|
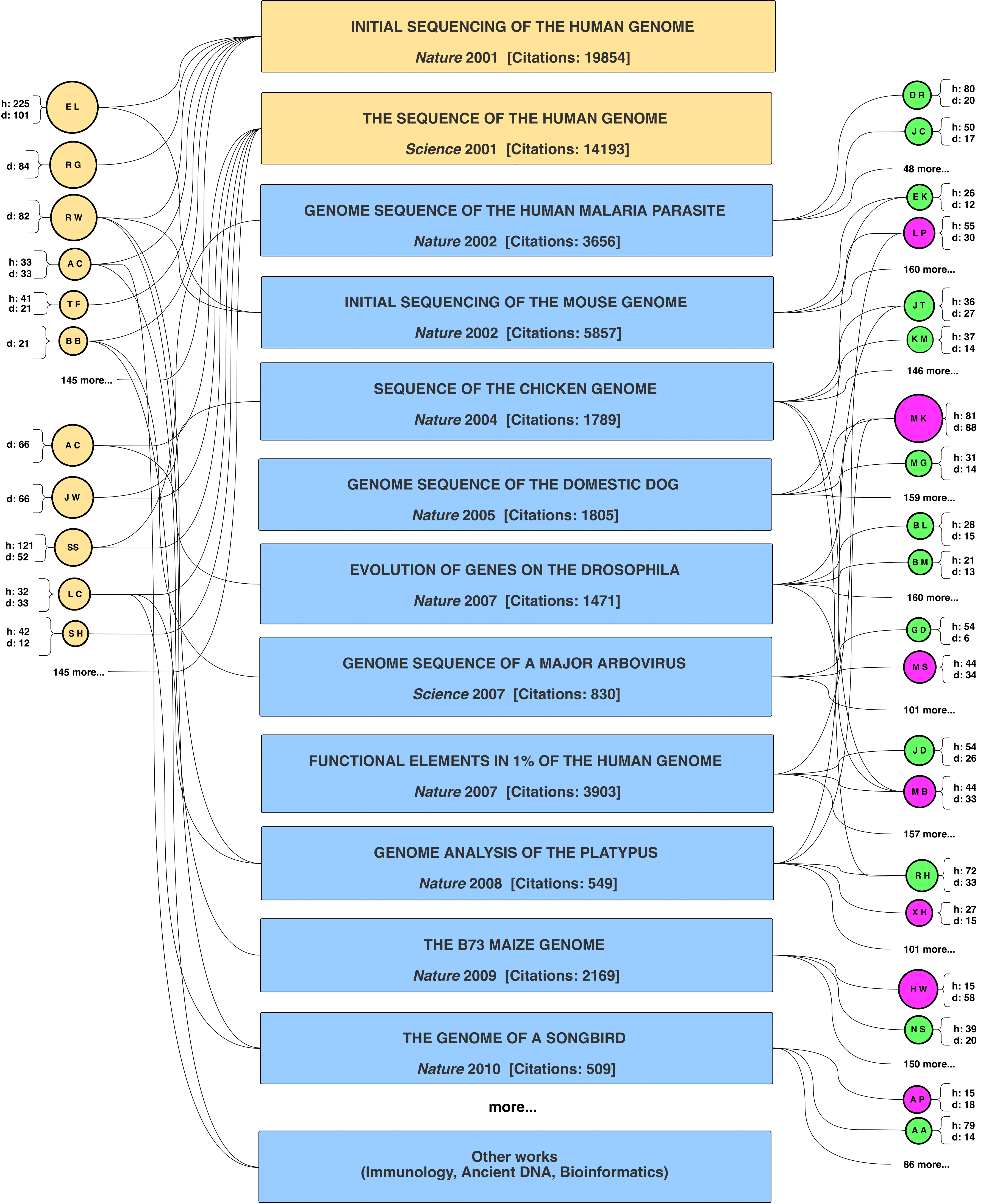
FIGURE 2: Evolution of the Genomics Revolution - the story behind the numbers Consortium-level collaborations of the Human Genome Project (HGP) scholars with faculty in biology and computing departments during the 2000s, and some of the key publications they produced, powering the genomics revolution. The scholar nodes bear the name initials. On the left panels, one can recognize some well-known HGP scholars, such as Eric Lander (EL) and Bruce Birren (BB). “d” stands for the network degree of a scholar, and is commensurate with the size of her/his node. “h” stands for the h-index of a scholar. Green nodes denote faculty affiliated with biology departments. Magenta nodes denote faculty affiliated with computing departments. Credit: The figure was generated by the panel model reported by Petersen et al., Sci. Adv. 2018;4: eaat4211 |
|
|
|
|
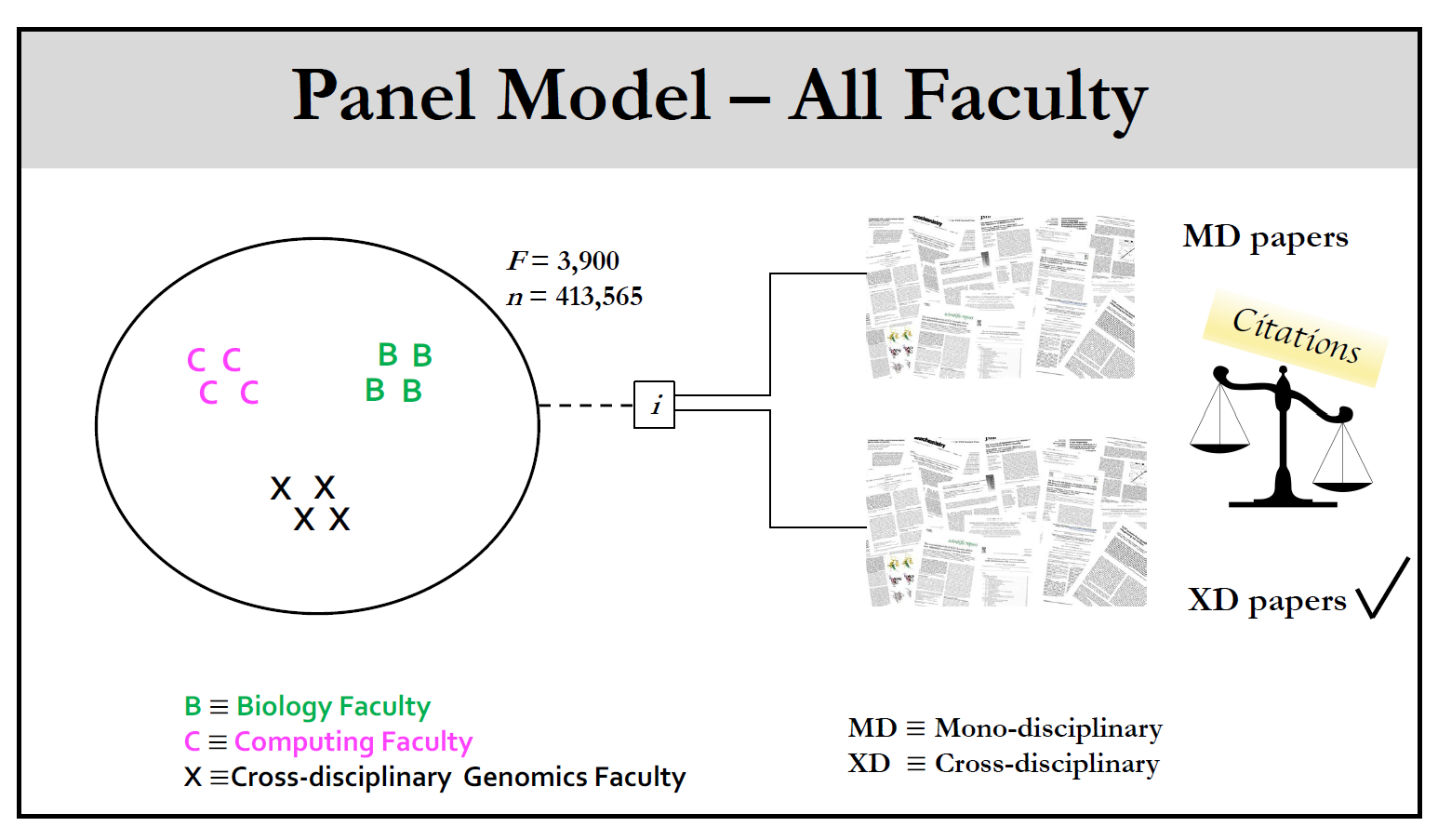
FIGURE 3: Panel analysis of scientific impact On average, cross-disciplinary faculty in genomics had more impactful careers with respect to their mono-disciplinary colleagues in biology and computing departments in the U.S. Career impact was measured in citations. The source of this higher impact has been traced to the cross-disciplinary portion of their publication portfolio. Credit: The figure was generated by the panel model reported by Petersen et al., Sci. Adv. 2018;4: eaat4211 |
|
|
|
Cartoon Art: Biologists working in tune with computer scientists propelled the genomics revolution Credit: Prof. Ergun Akleman, Texas A&M Visualization Department (crafted specifically for the paper Petersen et al., Sci. Adv. 2018;4: eaat4211) |
|
|
|
FUNDING: This research was supported by the National Science Foundation through grant 1738163, entitled “From Genomics to Brain Science.” 
INVESTIGATORS: Ioannis Pavlidis - Alexander M. Petersen
RESEARCH ASSISTANTS: Dinesh Majeti - Mohammed Emtiaz Ahmed
INSTITUTIONS: Computational Physiology Laboratory, University of Houston
Ernst and Julio Gallo Management Program, University of California, Merced
PAPER: To read the paper go to: Petersen et al., Sci. Adv. 2018;4: eaat4211
DATA: To access the dataset associated with the paper go to: https://osf.io/7nb6d/
BLOG: To read the story behind this research, access the blog of Dr. Pavlidis at: WordPress